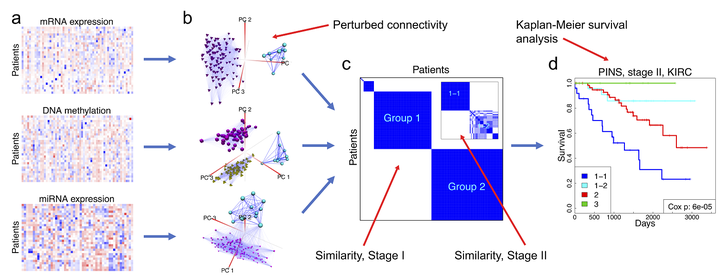
PINSPlus is fast and supports the analysis of large datasets with hundreds of thousands of samples and features. The software automatically determines the optimal number of clusters and then partitions the samples in a way such that the results are robust against noise and data perturbation. PINSPlus improves two algorithms of PINS (Nguyen et al., 2017; Nguyen, 2017; Nguyen et al., 2019): i) PerturbationClustering() to cluster a single data type, and ii) SubtypingOmicsData() to integrate omics data. When only a single data type is available, PINSPlus uses PerturbationClustering() function to perform sub-typing. The method is super fast, highly scalable, and can subtype hundreds of thousands of samples in under three minutes. We implement an ensemble strategy to optimize the running time while maintaining patient partitioning performance. We use singular value decomposition (SVD) and randomized singular value decomposition (RSVD) to project the original data to lower dimensional space. Then, we repeatedly perturb the subspace data by adding Gaussian noise and cluster the patients using different cluster numbers. The clustering assignment that gives the best agreement between the perturbed and original data yields the optimal subtype. When the dataset contains a large number of patients, we perform subtyping on a subset of size 2,000 and map the unpartitioned patients to the closest subtype using K nearest neighbors (KNN) algorithm. We use the SubtypingOmicsData() function when muti-omics data types are available. The method performs multiple stages of perturbation clustering and outputs patient connectivity graphs for each data type. The graphs that are resilient across all data types yield the most agreed number of patient subtypes.